Unlocking the secrets hidden in Next-Generation Sequencing (NGS) data is an exciting journey, but it’s important to ensure that the data quality is top-notch. That’s where powerful tools like FastQC come in! With FastQC, you can easily perform quality checks and ensure that your data is of the highest caliber. In this post, we delve into real-life cases where diagnostic plots from FastQC have helped unravel complex problems and bring clarity to our analysis. Join me on this thrilling adventure!
The arrangement of this post is based on the output from FastQC, but the criteria or cases mentioned here should also be applicable to similar measurements produced by other tools.
Per Base Sequence Content
Per Base Sequence Content stacks together all sequences in a fastq file and calculates the frequency of the four normal DNA bases (A/T/C/G) called for each base position. In a random or unbiased library, the bases should follow a random distribution of A/T/C/G, so the lines in the plot should be parallel to each other (around 25%). However, the actual distributions may have some fluctuations, depending on the overall number of bases in the genome and the capturing bias from the assay. Nevertheless, in most cases, the lines should be approximately parallel.
Per Base Sequence Content can be used to find:
- biased fragments, like:
- untrimmed barcodes. For demultiplexed libraries, if there are untrimmed barcodes, then because of the fixed sequences, you would observe sharp peaks at the beginnings or ends of reads. Below is an example showing that $5^\prime$ barcodes (TGGTCAC) are not trimmed:
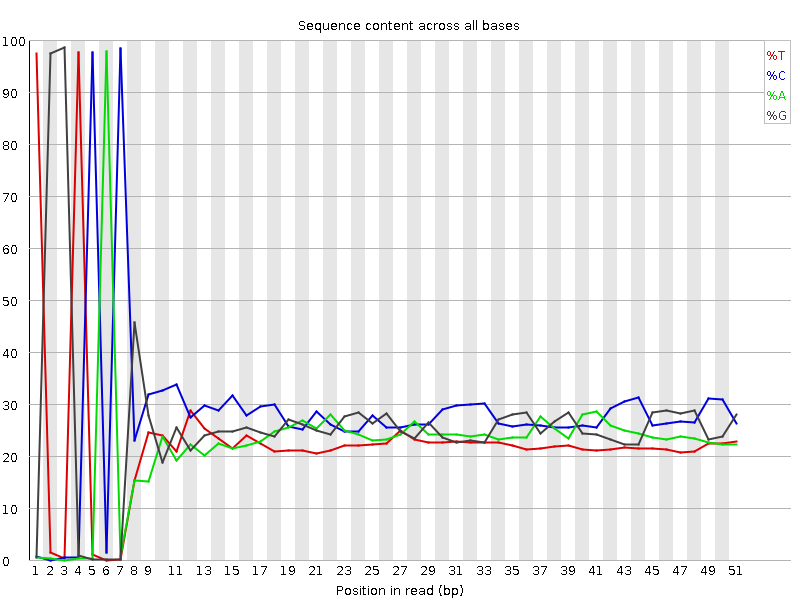
- template switching oligo. In cases like this, you can observe characteristic trinucleotide GGG or CCC near the beginning of reads.
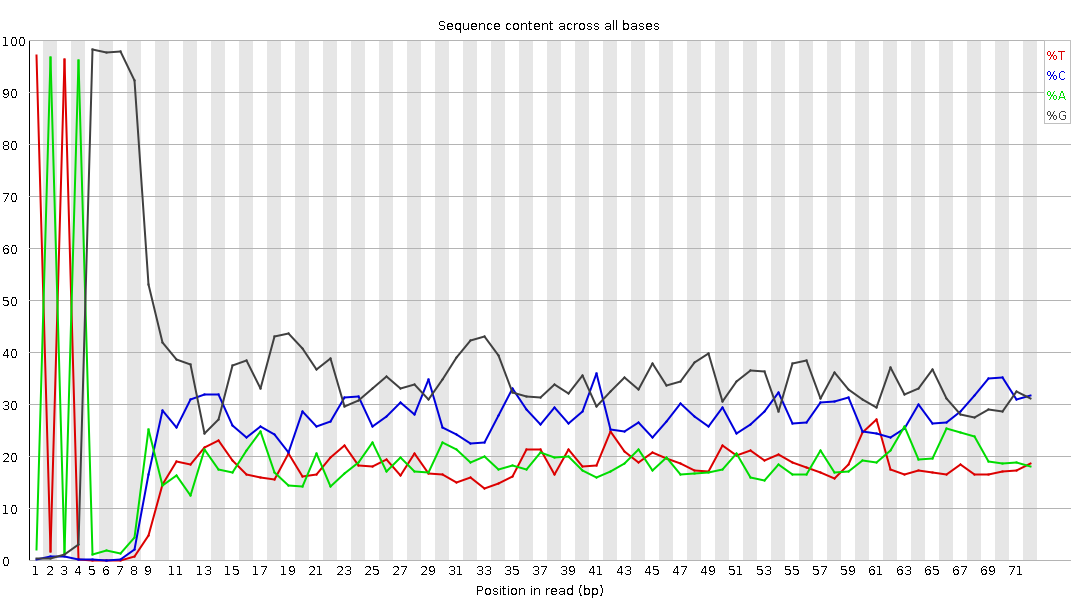
- untrimmed barcodes. For demultiplexed libraries, if there are untrimmed barcodes, then because of the fixed sequences, you would observe sharp peaks at the beginnings or ends of reads. Below is an example showing that $5^\prime$ barcodes (TGGTCAC) are not trimmed:
- overrepresented sequences, like adapter dimers or rRNAs.
Safelist:
- For libraries treated with sodium bisulfite, which will convert C to T, it’s normal to observe a low percent of Cs.
Adapter Content
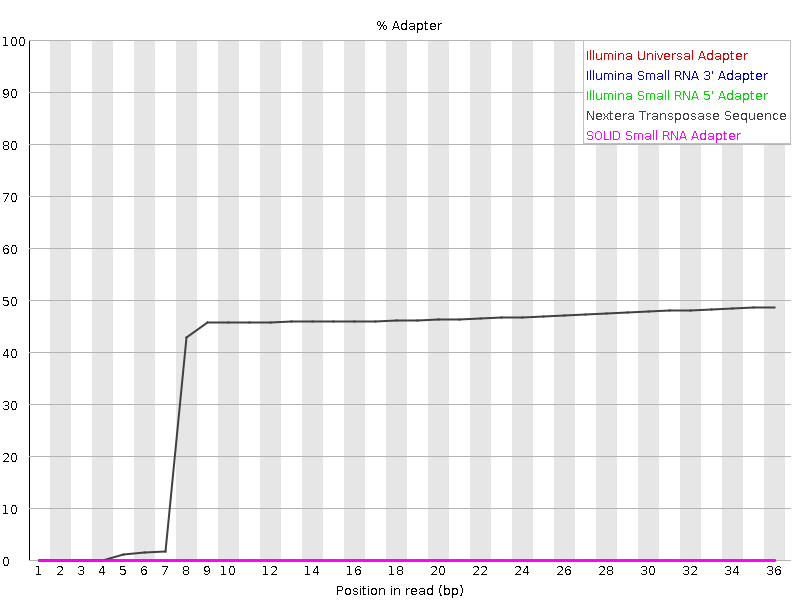
For libraries where a significant amount of the inserts are shorter than the sequencing length, adapters are likely to be incorporated in final reads. This is very common for libraries enriching for short/small RNAs, like PRO-cap, PRO-seq, etc. The Adapter Content module compares reads with commonly used adapter sequences and plots the enrichment. Adapter sequences may greatly affect on sequencing alignments, so if you see warnings in this section, you may need to trim adapters with cutadapt, fastp, or any other tool you like. In the following example, the Adapter Content module detects the existence of Nextera Transposase Sequence (the risen black line).
Overrepresented Sequences
A sequencing library typically consists of a diverse mixture of DNA or RNA molecules, and the presence of frequently occurring specific sequences can indicate an abnormality.
Possibility 1: Adapters
One possible cause for warnings or errors from this module is the presence of adapters. For example, people usually use a customized $3^\prime$ cloning adapter, CTGTAGGCACCATCAAT, to generate Ribo-seq libraries. This sequence is not included in the known-adapter list in FastQC, so the Adapter Content module cannot detect the existence of adapters, but the Overrepresented Sequences module catches a lot of hits. The following screenshot shows the top 12 overrepresented sequences from a Ribo-seq library (SRR942878), and the adapter sequence is highlighted for clarity.
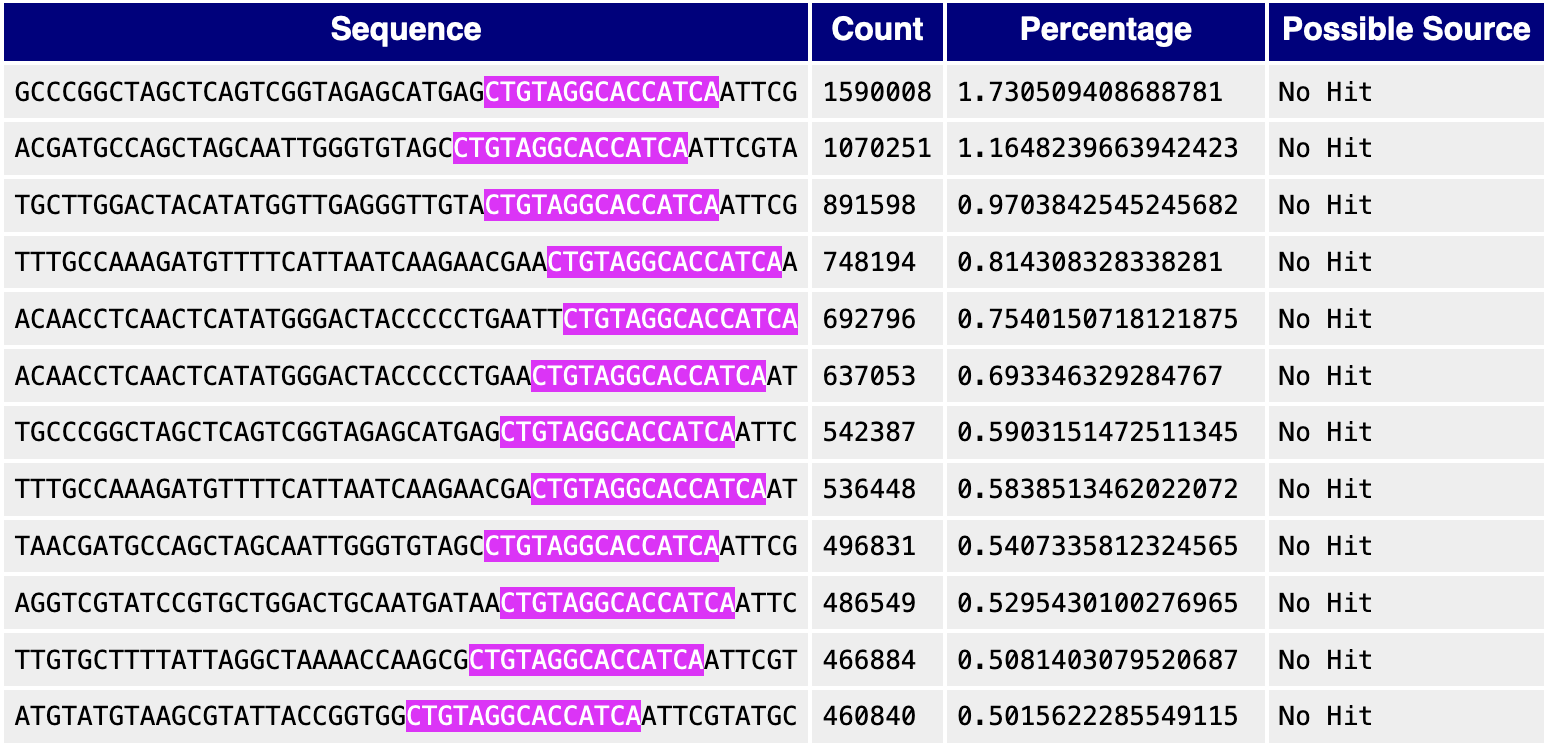
In practical applications, adapters may not be immediately visible in a table of many sequences. To identify potential adapters, I often choose a few overrepresented sequences and use the Smith-Waterman alignment algorithm to find adapter candidates. Typically, adapters appear as aligned contigs near the ends of sequences. For instance, when running the SW alignment on the first two hits in the table above, the adapter CTGTAGGCACCATCAAT is clearly visible:
1 | CCGGCTAGCTCAGTCGGTAGAGCATGAGCTGTAGGCACCATCAATTCG |
You can run the Smith-Waterman algorithm here.
Possibility 2: rRNAs or Other “Contamination”
What if the overrepresented sequences don’t match the typical adapter patterns? In some cases, these sequences may actually be “contamination” from ribosomal RNAs or other sources. One way to test this possibility is to run a BLAST search on the sequences. For example, here are the top overrepresented sequences for another sequencing library:
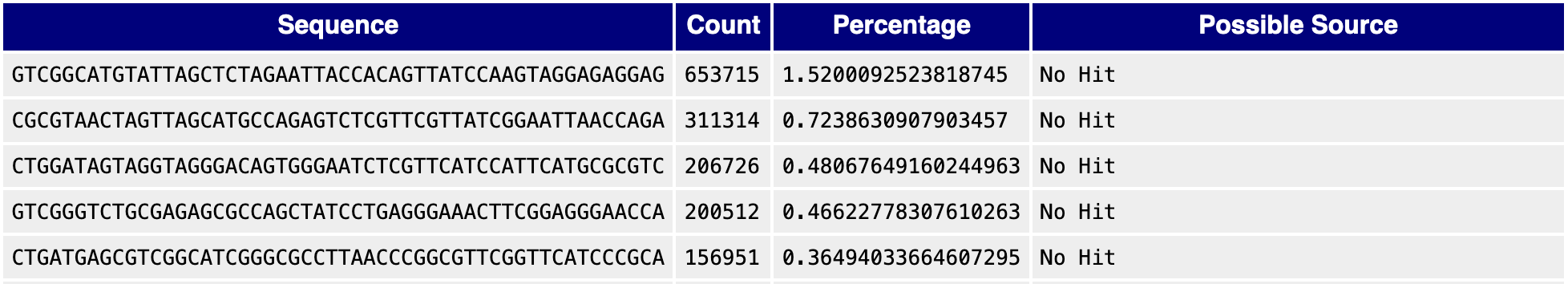
When running BLAST on some of these overrepresented sequences, you can see that they match with ribosomal RNAs.
1 | Chain 2, 18S rRNA |
If this is the case, you can align the entire library to ribosomal RNAs first, then do a second round of alignment with reads that cannot be aligned in the first round. This ensures you are left with high-quality reads for downstream analysis.
Per Base Sequence Quality
No cases yet.
Per Sequence Quality Scores
No cases yet.
Per Sequence GC Content
No cases yet.
Per Base N Content
No cases yet.
Sequence Length Distribution
No cases yet.
Sequence Duplication Levels
No cases yet.
Reference: FastQC manual